Building a Simple AI Conversational Assistant with Langchain.js
Table of Contents
For my final undergraduate professional course, Software Development Practicum, I was tasked with developing a virtual veterinary hospital learning system that required implementing an AI conversational assistant. The requirements were somewhat ambiguous. Considering that the other components consisted mainly of basic CRUD operations, my initial approach was to explore integrating these operations with an AI dialogue system capable of querying information about medications and clinical cases stored in the database.
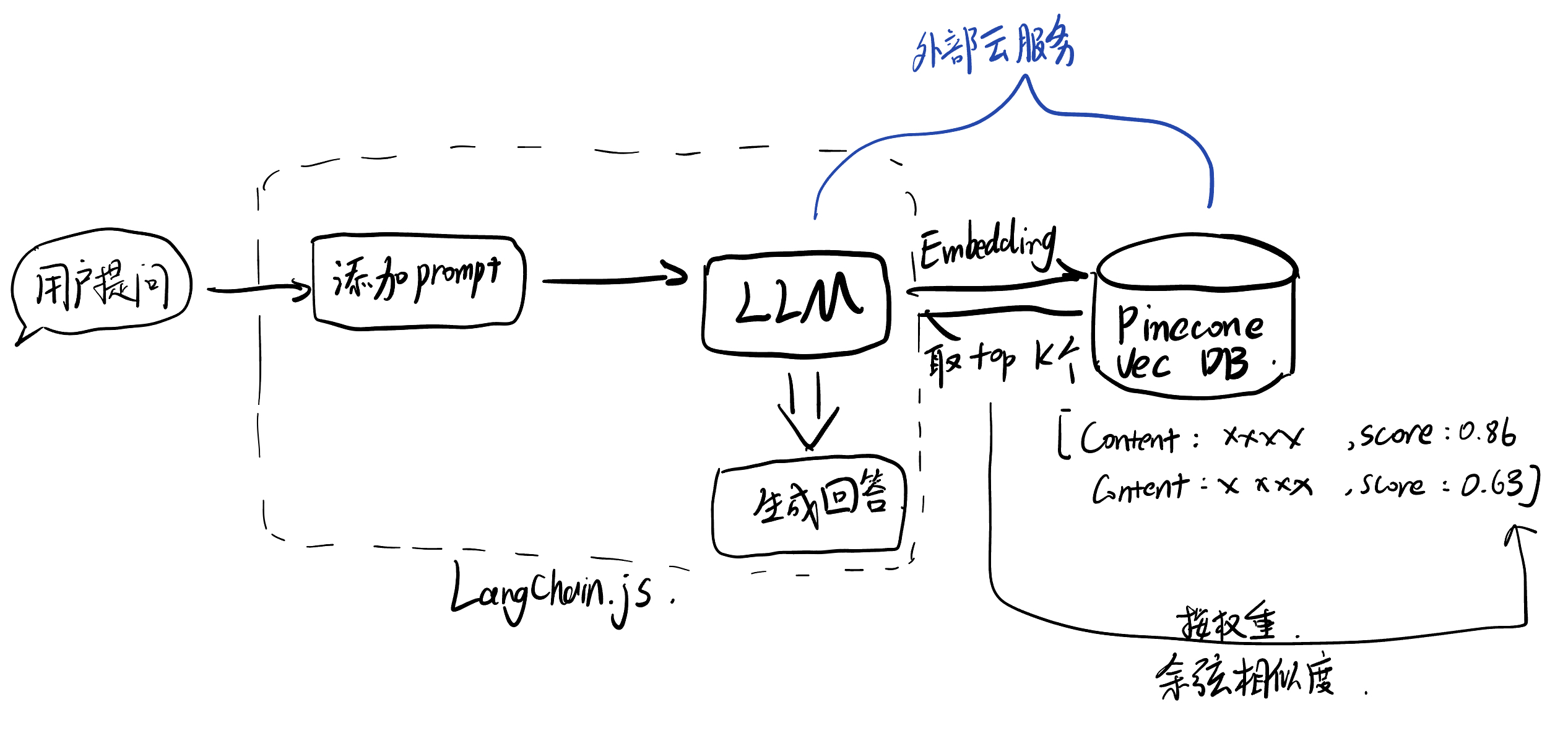
The architecture I adopted was a Retrieval-Augmented Generation (RAG) framework. Following the principle of simplicity, I implemented the solution using Langchain.js entirely on the frontend, utilizing OpenAI’s API for language processing and Pinecone’s remote vector database service.
What is RAG? (from GPT-4)
Retrieval-Augmented Generation (RAG) is a technique that combines information retrieval with generative models to enhance the quality and relevance of natural language processing tasks. It leverages external knowledge sources (such as document databases or information repositories) to augment the capabilities of generative models, making it particularly effective for question answering, summarization, translation, and similar tasks.
How RAG Works
A RAG model consists of two main components: retriever and generator.
- Retriever:
- The retriever’s task is to identify and extract the most relevant documents or information fragments from a large collection of documents in response to an input query. This is typically implemented through vector similarity search, where both queries and documents are encoded as vectors, and similarity metrics are used to find the best matches.
- Generator:
- The generator is typically a Transformer-based language model, such as GPT or BERT. It uses the retrieved documents as contextual information to generate answers. This allows the generated content to rely not only on the model’s internal knowledge (learned during training) but also on external, specific information directly relevant to the current query.
Advantages of RAG
- Enhanced Information Richness: Compared to traditional generative models, RAG increases the amount of information available for processing queries by accessing external databases.
- Improved Accuracy and Relevance: For tasks that depend on external knowledge, RAG can provide more accurate and detailed answers.
- Flexibility: The ability to access any form of structured or unstructured data sources as needed.
Application Scenarios
RAG models are highly effective in various applications, including:
- Question Answering Systems: Capable of generating detailed answers, especially when fact-checking through external information sources is required.
- Content Summarization: Extracting key information from related documents to generate concise summaries.
- Article Writing: Referencing and integrating various resources and reports when creating content, enhancing richness and depth.
Technical Challenges
Despite its numerous advantages, implementing RAG technology presents several challenges:
- Retrieval Efficiency: Efficient retrieval of relevant documents is critical for real-time applications, requiring optimization of indexing and query processing.
- Data Synchronization: Ensuring that the external knowledge base is up-to-date is crucial for guaranteeing the accuracy and relevance of generated content.
- Result Consistency: Generated content must maintain consistency with retrieved documents to avoid producing misleading or incorrect information.
Implementation Details
First, I registered for an account on the Pinecone website (steps omitted).
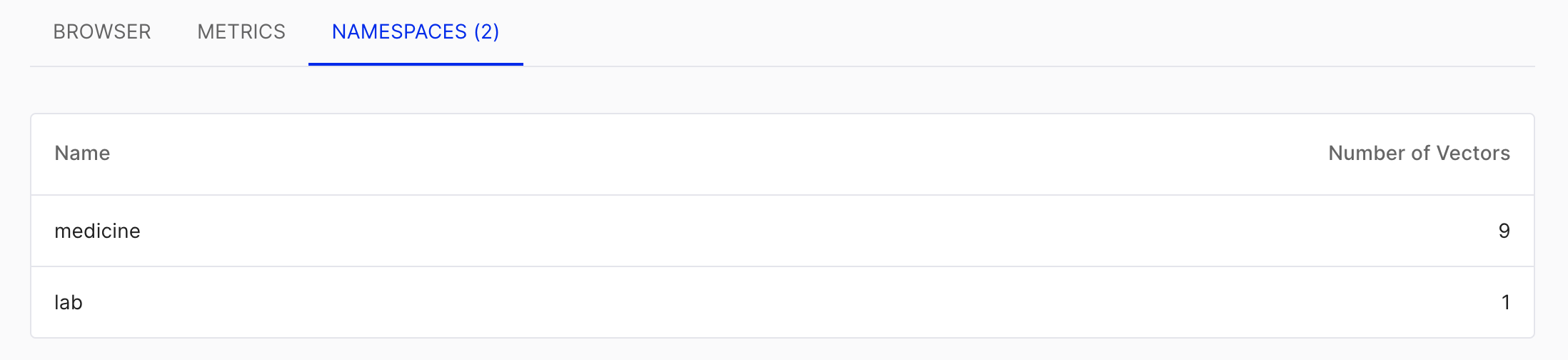
In Pinecone, a single project can contain multiple indices, and each index can be divided into multiple namespaces. Since I was using the free tier, which limits users to one project and one index, I differentiated between different sections using namespaces.
For instance, this project involves queries for medications (medicine) and laboratory tests (lab), so I defined these two namespaces accordingly. Namespaces are specified as parameters during insertion and don’t require manual setup. When querying, you can search within specific namespaces, which is quite convenient.
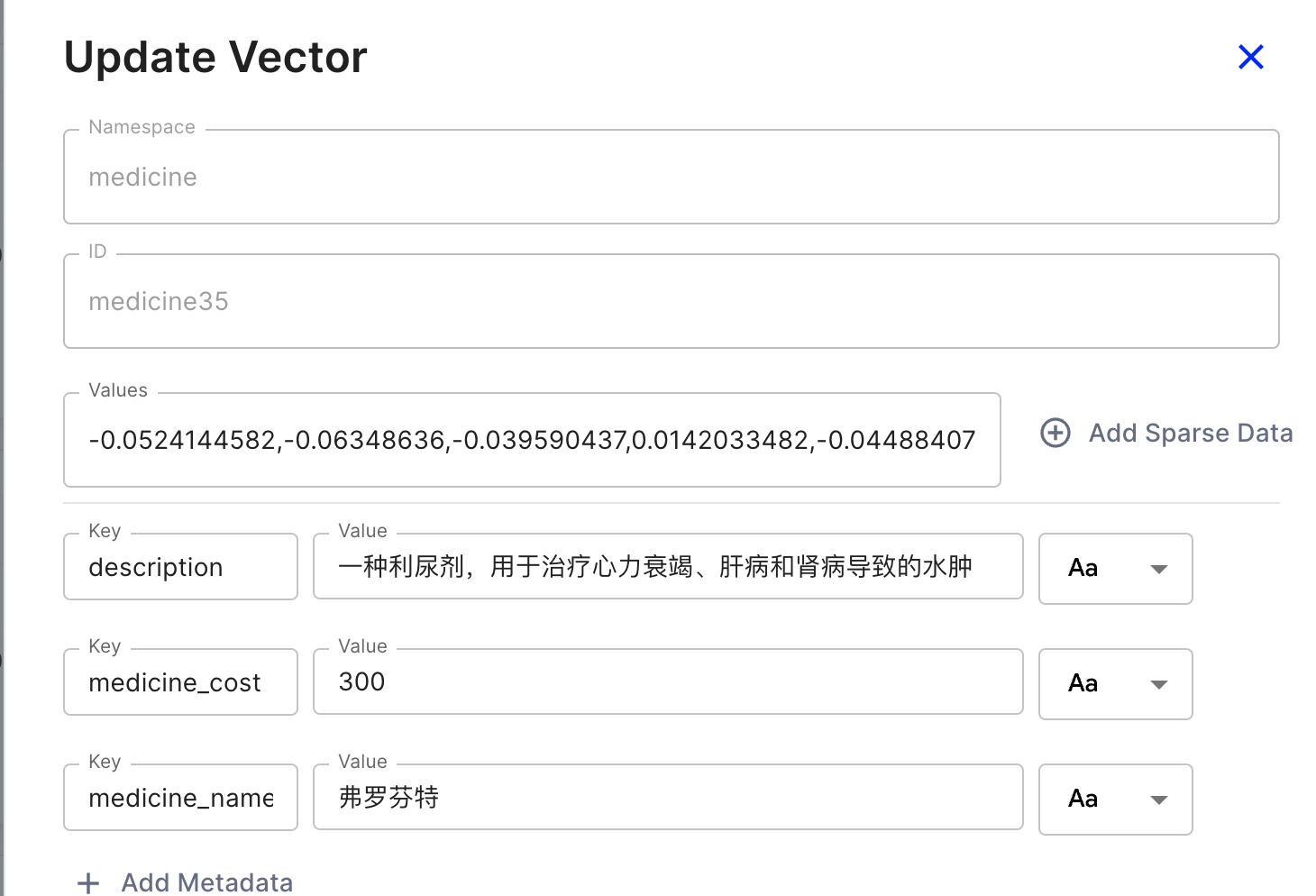
For each record, the namespace and ID are immutable, but you can update the Values and metadata (the Key-Value pairs shown below) based on the ID.
For the Values, these are arrays converted during the embedding process. I used OpenAI’s text-embedding-ada-002 model on the frontend, which produces 1536-dimensional vectors, hence the length of 1536.
export const pineconeAdd = async (id, namespace, input_text, metadata) => {
const embedding = await embedText(input_text);
try {
const insertResponse = await index.namespace(namespace).upsert([
{
id: `${namespace}${id}`,
values: embedding,
metadata: metadata
}
]);
return { success: true, message: 'Pinecone insertion successful', detail: insertResponse };
} catch (error) {
return { success: false, message: 'Pinecone insertion error', error };
}
};
The logic for the frontend insertion part is executed simultaneously with the insertion into MySQL. The update process is similar:
// Insert data into Pinecone
if (newMedicine.value.saveToPinecone) {
const input_text = "Medicine Name: " + newMedicine.value.medicine_name + ", Medicine Cost: "+ newMedicine.value.medicine_cost.toString() + ", Efficacy and Usage: " + newMedicine.value.description
const insert_pinecone = await pineconeAdd(
medicine_id,
`medicine`, input_text,
{
medicine_name: newMedicine.value.medicine_name,
medicine_cost: newMedicine.value.medicine_cost,
description: newMedicine.value.description
}
)
if (insert_pinecone?.success){
ElMessage({
message: 'Pinecone insertion successful',
type: 'success',
});
}else{
ElMessage.error(`Pinecone insertion failed: ${insert_pinecone}`);
}
}
Logic for handling the AI page:
if (queryType.value === 'medicine') {
const queryResponse = await pineconeIndex.namespace('medicine').query({
topK: 5,
vector: await embedText(userInput.value),
includeMetadata: true,
})
console.log("Query Response:", queryResponse);
responseText.value = "Querying...";
if (queryResponse?.data?.length === 0) {
responseText.value = "No relevant information found";
return;
}
const formattedData = medicineFormatForLLM(queryResponse);
console.log("Formatted Data for LLM:", formattedData);
const llmQuery = getMedicineLLMQuery(formattedData, userInput.value);
console.log("LLM Query:", llmQuery);
responseText.value = llmQuery;
console.log(userInput.value);
const response = await chat.invoke([
new HumanMessage(
llmQuery
),
]);
console.log(response);
responseText.value = response.content as string;
}
Utility functions and the delete function used:
export const medicineFormatForLLM = (queryResponse) => {
return queryResponse.matches.map(match => ({
id: match.id,
name: match.metadata.medicine_name,
cost: match.metadata.medicine_cost,
description: match.metadata.description,
score: match.score
}));
};
export const getMedicineLLMQuery = (data, userInput) => {
// Convert data to text format for use as query context
const context = data.map(item => `ID: ${item.id}, Medicine Name: ${item.name},
Medicine Cost (in RMB): ${item.cost}, Description: ${item.description}`).join('\n');
return `Based on the following retrieved medicine information:\n${context}\nAnswer the user's question in Chinese:\n${userInput}.`;
};
export const pineconeDelete = async (id, namespace) => {
const ns = index.namespace('medicine');
await ns.deleteOne(`${namespace}${id}`);
}
Text retrieval involves first vectorizing the user’s query, then comparing it with the values using cosine similarity, extracting the top K most similar texts, and generating a response by combining the texts with the user’s query and a custom prompt.
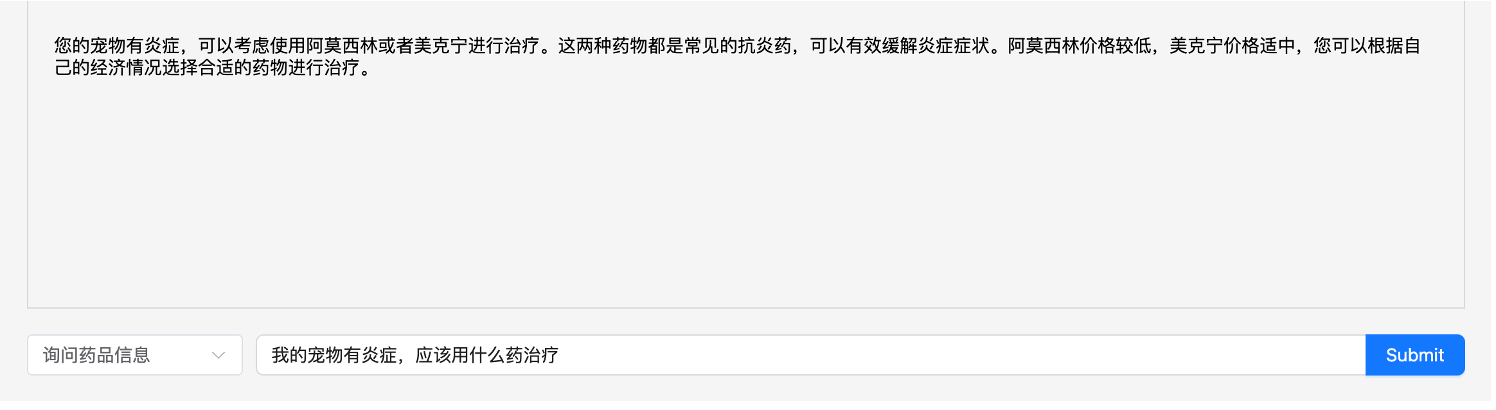
Results:
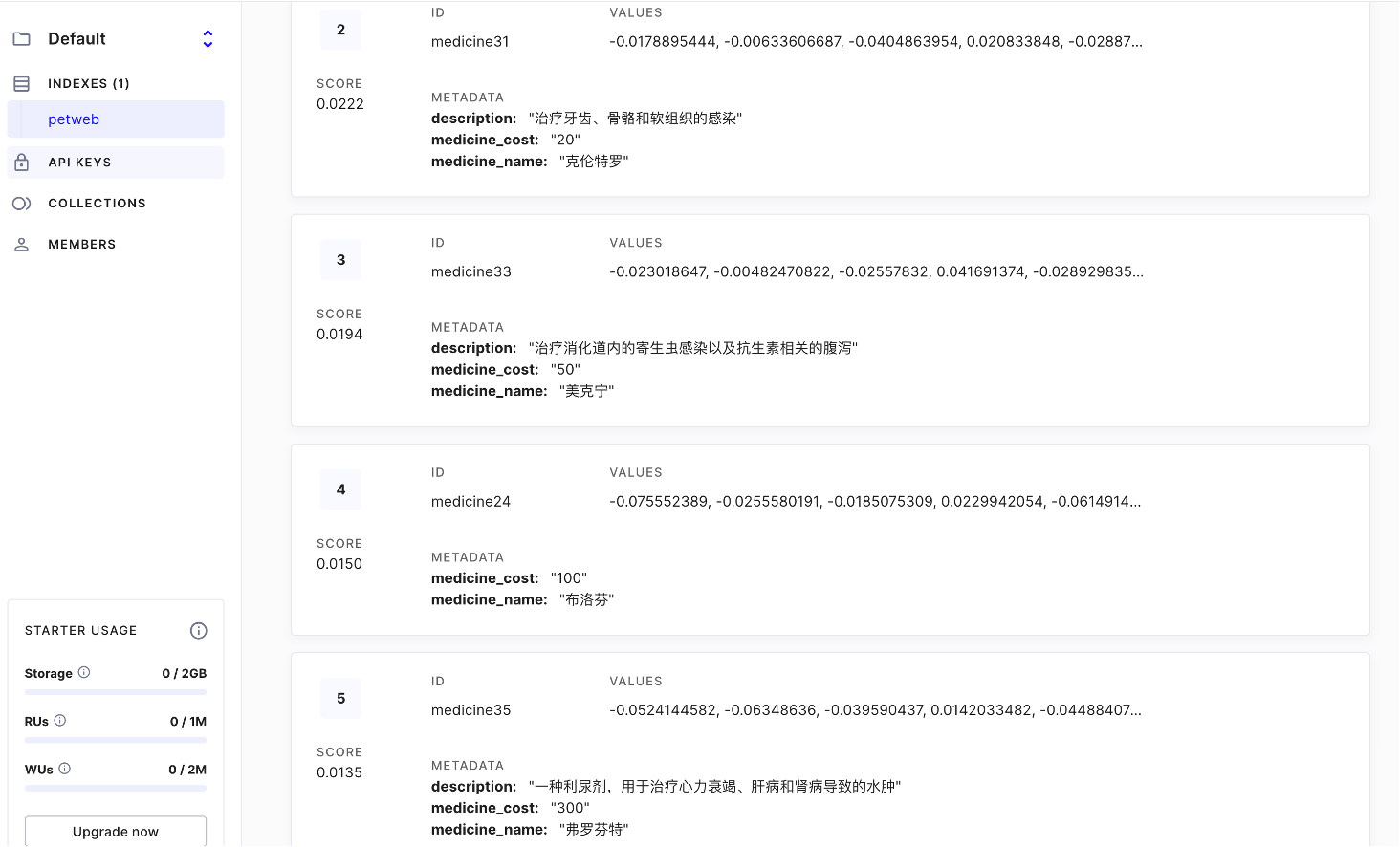
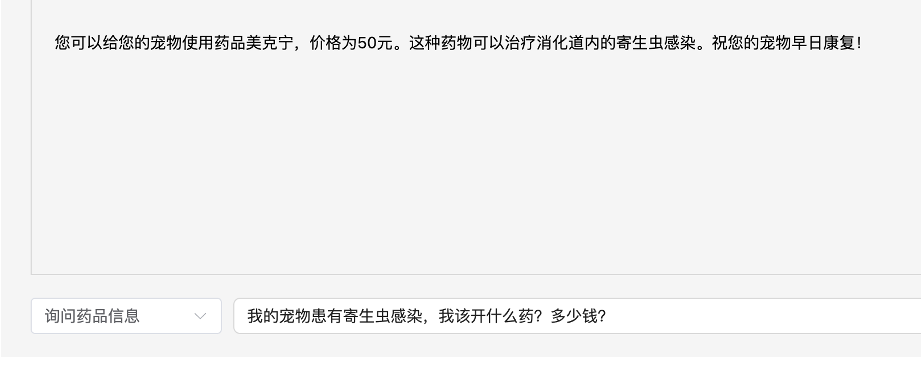
Finally, a note on Langchain: the JavaScript version and documentation are quite lacking. When I used the Python version for my thesis, I used a vectorStore, but following the documentation for the JavaScript version, it was completely unusable. In the end, I only used some basic functions, which seemed almost unnecessary.